Resampling
Resampling#
The resampling step is the most crucial task of the preprocessing. Using the samples from the preparation step, the resampling stitches together the used processes and, based on the resampling method used, resamples all flavours used to one target flavour. Due to differences in the number of jets in different p_T and \eta regions, the tagging of the jets is not independent of those regions. To ensure a kinematic independent tagging of the jets, the resampling methods sample the different flavours so that in the given p_T and \eta bins, the same amount of jets per flavour are present. The technique how this is done are specific to the method. In their respective section, this will be explained more in detail. This 2D resampling is illustrated with the following plot from Manuel Guth's PhD Defense Talk:
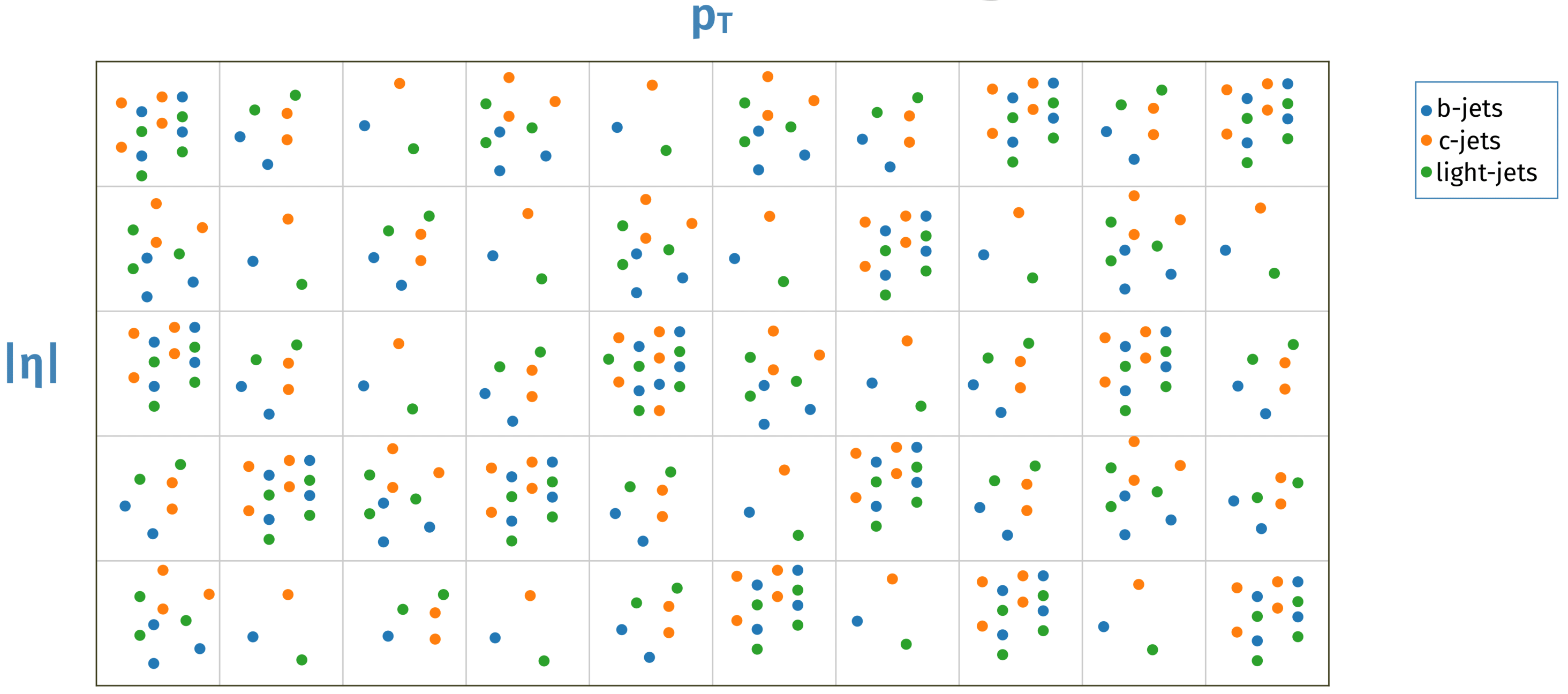
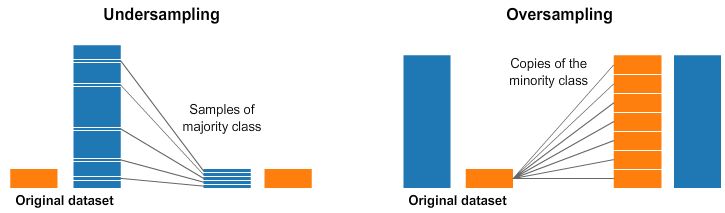
For the count (or down-sampling), PDF (a combination fo up- and down-sampling) and importance_no_replace (similar to PDF but no reusing of the jets) methods, we first fill the target flavour in your 2D bins. After that, for each bin, the same amount of jets from other flavours, which fit in the 2D bin, are added. This up- and down-sampling is illustrated in the following plot from Kaggle:
The Weighting Sampling is not a real resampling, in the same sense as the others. To achieve the kinematic independent training sample, weights are calculated for all jets according to their p_T and \eta values. This is similar to a 2D reweighting.
General Config File Options#
The resampling part of the config file starts with the sampling: dict.
sampling:
# Classes which are used in the resampling. Order is important.
# The order needs to be the same as in the training config!
class_labels: [ujets, cjets, bjets]
# Decide, which resampling method is used.
method: count
In sampling, we can define now the method which is used in the preprocessing for resampling. method defines the method which is used. Currently available are count, pdf, importance_no_replace and weighting. The details of the different sampling methods are explained at their respective sections. The here shown config is for the count method.
An important part are the class_labels which are defined here. You can define which flavours are used in the preprocessing. The name of the available flavours can be found here. Add the names of those to the list to add them to the preprocessing. PLEASE KEEP THE ORDERING CONSTANT! THIS IS VERY IMPORTANT. This list must be the same as the one in the train config!
For an explanation of the resampling function specific options, have a look in the section of the resampling method you want to use. The general options are explained in the following:
# Fractions of ttbar/zprime jets in final training set. This needs to add up to one.
fractions:
ttbar: 0.7
zprime: 0.3
# number of training jets
# For PDF sampling: the number of target jets per class!
# So if you set n_jets=1_000_000 and you have 3 output classes
# you will end up with 3_000_000 jets
# For other sampling methods: total number of jets after resampling
# If set to -1: max out to target numbers (limited by fractions ratio)
n_jets: 25e6
# number of validation jets in the hybrid validation sample
# Same rules as above for n_jets when it comes to PDF sampling
n_jets_validation: 4e6
# Bool, if track information (for DIPS etc.) are saved.
save_tracks: True
# Name(s) of the track collection(s) to use.
tracks_names: ["tracks"]
# Bool, if track labels are processed
save_track_labels: True
# this stores the indices per sample into an intermediate file
intermediate_index_file: *intermediate_index_file
# this stores the indices per sample for the hybrid validation
intermediate_index_file_validation: *intermediate_index_file_validation
# for method: weighting
# relative to which distribution the weights should be calculated
weighting_target_flavour: 'bjets'
# If you want to attach weights to the final files
bool_attach_sample_weights: False
# How many jets you want to use for the plotting of the results
# Give null (the yaml None) if you don't want to plot them
n_jets_to_plot: 3e4
| Setting | Type | Explanation |
|---|---|---|
fractions |
dict |
Gives the fractions of t\bar{t} and Z' in the final training sample. These values need to add up to 1! |
n_jets |
int |
Number of target jets to be taken. For PDF sampling, this is the number of jets per class in the final training sample, while for other methods it is the total number of jets after resampling. For the pdf method, setting this value to -1 maximises the number of jets in the training sample. Note: The -1 option is only available for the pdf method! |
save_tracks |
bool |
Define if tracks are processed or not. These are not needed to train DL1r/DL1d |
tracks_names |
list of str |
Name of the tracks (in the .h5 files coming from the dumper) which are processed. Multiple tracks datasets can be preprocessed simultaneously when two str are given in the list. |
save_track_labels |
bool |
If this value is True, the track variables in track_truth_variables will be processed as labels without scaling. The will be saved in an extra group in the final training file. The name will be Y_<track_name>_train. <track_name> is here the name of the track collection. |
track_truth_variables |
str or list |
Track variables that will be handled as truth labels. Multiple can be given in a list of str or just one in a single string. |
intermediate_index_file |
str |
For the resampling, the indices of the jets to use are saved in an intermediate indices .h5 file. You can define a name and path in the Preprocessing-parameters.yaml. |
n_jets_scaling |
int |
Number of jets which are used to calculate scaling and shifting values. If null is given, all jets in file are used. |
n_jets_to_plot |
int |
Number of jets which are used for plotting the variables of the jets/tracks after each preprocessing step (resampling, scaling, shuffling/writing). If null is given, the plotting is skipped. |
Note: n_jets are the number of jets you want to have in your final training file for the count and weighting method. For the pdf method, this is the number of jets per flavour in the training file!
Count Sampling#
Standard undersampling approach. Undersamples all flavours to the statistically lowest flavour used.
sampling_variables:
- pt_btagJes:
# bins take either a list containing the np.linspace arguments
# or a list of them
# For PDF sampling: must be the np.linspace arguments.
# - list of list, one list for each category (in samples)
# - define the region of each category.
bins: [[0, 600000, 351], [650000, 6000000, 84]]
- absEta_btagJes:
# For PDF sampling: same structure as in pt_btagJes.
bins: [0, 2.5, 10]
# Decide, which of the in preparation defined samples are used in the resampling.
samples_training:
ttbar:
- training_ttbar_bjets
- training_ttbar_cjets
- training_ttbar_ujets
zprime:
- training_zprime_bjets
- training_zprime_cjets
- training_zprime_ujets
# Decide, which of the in preparation defined samples are used in the hybrid
# validation resampling.
samples_validation:
ttbar:
- validation_ttbar_bjets
- validation_ttbar_cjets
- validation_ttbar_ujets
zprime:
- validation_zprime_bjets
- validation_zprime_cjets
- validation_zprime_ujets
custom_n_jets_initial:
# these are empiric values ensuring a smooth hybrid sample.
# These values are retrieved for a hybrid ttbar + zprime sample for the count method!
training_ttbar_bjets: 5.5e6
training_ttbar_cjets: 11.5e6
training_ttbar_ujets: 13.5e6
# These are the values for the hybrid validation sample. Just use the same as above!
validation_ttbar_bjets: 5.5e6
validation_ttbar_cjets: 11.5e6
validation_ttbar_ujets: 13.5e6
| Setting | Type | Explanation |
|---|---|---|
sampling_variables |
list |
Needs exactly 2 variables. Sampling variables which are used for resampling. The example shows this for the pt_btagJes and absEta_btagJes variables. In case of the count method, you define a nested list (one sublist for each category (t\bar{t} or Z')) with the first and last bin edge and the number of bins to use. |
custom_n_jets_initial |
dict |
Used jets per sample to ensure a smooth hybrid sample of t\bar{t} and Z', we need to define some empirically derived values for the t\bar{t} samples. |
samples_training |
dict |
You need to define them for both of your samples. By default (if you want to use the standard hybrid of t\bar{t} and Z', they are named ttbar and zprime but they are just arbitrary names. The samples defined in here are the ones we prepared in the step above which will be used for the training sample. The samples defined in here are the ones we prepared in the step above which will be used for the training sample. To ensure a smooth hybrid sample of t\bar{t} and Z', we need to define some empirically derived values for the t\bar{t} samples in custom_n_jets_initial. |
samples_validation |
dict |
Same as the samples_training option but for when you want to produce the hybrid validation sample. |
Importance Sampling With Replacement (PDF Sampling)#
The PDF sampling method is based on the principles of importance sampling. If your sample's statistics are small and/or your lowest distribution is other than the target distribution (in case of b-tagging, this is the b-jet distribution), you can force the b-jet distribution shape on the other jet flavour distributions. This will ensure all the distributions have the target distribution shape and the same fractions for the two given resampling variables. To enforce the same shape and number of jets per p_T and \eta bin, the statistically higher flavours are undersampled and the statistically lower flavours are upsampled to the target flavour. An example for the reprocessing config file which uses the pdf sampling can be found here. In this case, four different flavours are used.
The options for the pdf method seems quite similar to the ones from the count method. But there are some important differences!
First are the bins for the two resampling variables. You need to define a nested list with the regions for both sample categories (t\bar{t} and Z'). Even if they are the same!
options:
sampling_variables:
- pt_btagJes:
# bins take either a list containing the np.linspace arguments
# or a list of them
# For PDF sampling: must be the np.linspace arguments.
# - list of of list, one list for each category (in samples)
# - define the region of each category.
bins: [[0, 25e4, 100], [25e4, 6e6, 100]]
- absEta_btagJes:
bins: [[0, 2.5, 10], [0, 2.5, 10]]
# Decide, which of the in preparation defined samples are used in the resampling.
samples_training:
ttbar:
- training_ttbar_bjets
- training_ttbar_cjets
- training_ttbar_ujets
zprime:
- training_zprime_bjets
- training_zprime_cjets
- training_zprime_ujets
# Decide, which of the in preparation defined samples are used in the hybrid
# validation resampling.
samples_validation:
ttbar:
- validation_ttbar_bjets
- validation_ttbar_cjets
- validation_ttbar_ujets
zprime:
- validation_zprime_bjets
- validation_zprime_cjets
- validation_zprime_ujets
custom_n_jets_initial: # Leave empty for pdf method
# For PDF sampling, this is the maximum upsampling rate (important to limit tau upsampling)
# File are referred by their key (as in custom_n_jets_initial)
max_upsampling_ratio:
training_ttbar_cjets: 5
training_zprime_cjets: 5
validation_ttbar_cjets: 5
validation_zprime_cjets: 5
# For PDF sampling, this scales the total number of training jets in the training dataset by
# given factor, i.e. a factor of 1 has no effect
sampling_fraction:
training_ttbar_cjets: 1
validation_ttbar_cjets: 1
| Setting | Type | Explanation |
|---|---|---|
sampling_variables |
list |
Needs exactly 2 variables. Sampling variables which are used for resampling. The example shows this for the pt_btagJes and absEta_btagJes variables. In case of the pdf method, you define a nested list (one sublist for each category (t\bar{t} or Z')) with the first and last bin edge and the number of bins to use (np.linespace arguments). |
custom_n_jets_initial |
None |
These values are used only in the count and weighting method. |
samples_training |
dict |
You need to define them for both of your samples. By default (if you want to use the standard hybrid of t\bar{t} and Z', they are named ttbar and zprime but they are just arbitrary names. The samples defined in here are the ones we prepared in the step above which will be used for the training sample. |
samples_validation |
dict |
Same as the samples_training option but for when you want to produce the hybrid validation sample. |
max_upsampling_ratio |
dict |
Here you can define for the different samples, which are defined in the samples section, a maximal ratio of upsampling. If there are not enough cjets and the max_upsampling_ratio is reached, the form of the distribution is applied but not the number. So there can be different numbers of jets per bin per class, but the shape of distributions will still be the same (if you normalise them). |
sampling_fraction |
dict |
Here you can define for the different samples, which are defined in the samples section, a factor to scale the number of jets for this sample in the final training dataset compared to the number of jets defined in n_jets. This can be useful if subclasses of u-, c- and/or b-jets are used for training but the overall ratio for u-,c- and b-jet should still be 1:1:1 |
Importance Sampling Without Replacement#
Method based on the principles of importance sampling. This method is similar to the count method but with the added flexibility of being able to take a target distribution which all the other distributions should fall under. The implementation also ensuring same fractions per flavor. The difference between this method and the PDF sampling method, is that examples/events are not repeated. You can force the b-jet distribution shape on the other jet flavour distributions by specifying the target distribution to be the b-jets. This will ensure all the distributions have the b-jets shape and same fractions for the two given resampling variables p_T and \eta . To enforce the same shape and number of jets per p_T and \eta bin, first the sampling probabilityies are calculated using target / distribution_i, where distribution_i is for each flavour, then the distributions are scaled up/down using the maximum sampling probability. The statistically higher flavours are undersampled and the statistically lower flavours are first scaled then downsampled to the target flavour.
The options for the this method are similar to the ones from the count method.
sampling:
# Downsampling method that gives same fractions and shape
# distributions given a target distribution, here the b-jets
method: importance_no_replace
options:
# Specify the target distribution
target_distribution: bjets
# jet variables used
sampling_variables:
- pt_btagJes:
# bins take either a list containing the np.linspace arguments
# or a list of them
bins: bins: [0, 15e5, 250]
- absEta_btagJes:
bins: [0, 2.5, 9]
# Decide, which of the in preparation defined samples are used in the resampling.
samples_training:
ttbar:
- training_ttbar_bjets
- training_ttbar_cjets
- training_ttbar_ujets
zprime:
- training_zprime_bjets
- training_zprime_cjets
- training_zprime_ujets
# Decide, which of the in preparation defined samples are used in the hybrid
# validation resampling.
samples_validation:
ttbar:
- validation_ttbar_bjets
- validation_ttbar_cjets
- validation_ttbar_ujets
zprime:
- validation_zprime_bjets
- validation_zprime_cjets
- validation_zprime_ujets
# Set to -1 or don't include this to use all the available jets
n_jets: -1
| Setting | Type | Explanation |
|---|---|---|
sampling_variables |
list |
Needs exactly 2 variables. Sampling variables which are used for resampling. The example shows this for the pt_btagJes and absEta_btagJes variables. In case of the pdf method, you define a nested list (one sublist for each category (t\bar{t} or Z')) with the first and last bin edge and the number of bins to use (np.linespace arguments). |
samples_training |
dict |
You need to define them for both of your samples. By default (if you want to use the standard hybrid of t\bar{t} and Z', they are named ttbar and zprime but they are just arbitrary names. The samples defined in here are the ones we prepared in the step above which will be used for the training sample. |
samples_validation |
dict |
Same as the samples_training option but for when you want to produce the hybrid validation sample. |
target_distribution |
str |
Target distribution to be used for computing the sampling probabilities relative to. This ensures all the final resampled distributions have the same shape and fraction as the target distribution. Default is the bjets. |
Weighting Sampling#
Alternatively you can calculate weights between the flavor of bins in the 2d(pt,eta) histogram and write out all jets. These weights can be forwarded to the training to weigh the loss function of the training. If you want to use them don't forget to set bool_attach_sample_weights to True.
options:
sampling_variables:
- pt_btagJes:
# bins take either a list containing the np.linspace arguments
# or a list of them
# For PDF sampling: must be the np.linspace arguments.
# - list of list, one list for each category (in samples)
# - define the region of each category.
bins: [[0, 600000, 351], [650000, 6000000, 84]]
- absEta_btagJes:
# For PDF sampling: same structure as in pt_btagJes.
bins: [0, 2.5, 10]
# Decide, which of the in preparation defined samples are used in the resampling.
samples_training:
ttbar:
- training_ttbar_bjets
- training_ttbar_cjets
- training_ttbar_ujets
zprime:
- training_zprime_bjets
- training_zprime_cjets
- training_zprime_ujets
# Decide, which of the in preparation defined samples are used in the hybrid
# validation resampling.
samples_validation:
ttbar:
- validation_ttbar_bjets
- validation_ttbar_cjets
- validation_ttbar_ujets
zprime:
- validation_zprime_bjets
- validation_zprime_cjets
- validation_zprime_ujets
# for method: weighting
# relative to which distribution the weights should be calculated
weighting_target_flavour: 'bjets'
# If you want to attach weights to the final files
bool_attach_sample_weights: False
| Setting | Type | Explanation |
|---|---|---|
samples_training |
dict |
You need to define them for ttbar and zprime. The samples defined in here are the ones we prepared in the step above which will be used for the training sample. |
samples_validation |
dict |
Same as the samples_training option but for when you want to produce the hybrid validation sample. |
weighting_target_flavour |
str |
To which distribution the weights are relatively calculated to. |
bool_attach_sample_weights |
bool |
Decide, if you want to attach these weights in the final training config. For all other resampling methods, this should be False. |
Running the Resampling#
The resampling can be started after the preparation step is finished with the command:
preprocessing.py --config <path to config file> --resampling
Parallel processing for the PDF method
If you want to also use the tracks of the jets, you need to set the option save_tracks in the preprocessing config to True. If the tracks have a different name than "tracks" in the .h5 files coming from the dumper, you can also set change tracks_names to your needs. Track information are not needed for the DL1r but for DIPS and Umami.
If you are using the pdf resampling method, you can further split this up into sub-components. First you need to start with working on the target distribution:
preprocessing.py --config <path to config file> --resampling --flavour target
This will process the target distribution for both categories, t\bar{t} and Z'. After this is finished, you can run in parallel the following step:
preprocessing.py --config <path to config file> --resampling --flavour $0
where the $0 stands for an index in samples/ttbar. If we take the example from the PDF Sampling section, this contains the samples of the bjets, cjets and ujets. 0 is therefore the bjets, 1 is the cjets and 2 is the ujets. Note: This will process the bjets for both categories, t\bar{t} and Z', although we need to look in samples/ttbar. Only the flavour is important here.
After all these subjobs are finished, you can continue with the plotting (this step plots the distributions of the resampled variables before the actual resampling) and the combination of the flavours into our final resampled file. For that you need to run
preprocessing.py --config <path to config file> --resampling --flavour plotting
and
preprocessing.py --config <path to config file> --resampling --flavour combining
where the plotting step can also be skipped if you want to.
Note: The steps defined in here are only performed on the training samples! You do not need to resample the validation/test samples execept you want to also produce the hybrid validation and test samples. For instructions to do that, please look at ADD ME.
Create the Resampled Hybrid Validation Sample#
To properly validate our training and to check against any issue while training (e.g. overfitting), we need a validation sample that is similar resampled as the training sample. When we use our non-resampled validation samples for that, we can't properly check the training behavior due to the difference in composition between the resampled training sample and the non-resampled validation sample(s).
To create the hybrid validation sample, you need to define the different flavours in the Preprocessing-samples.yaml (in the example, this is already done. They are called validation_<process>_<flavour>) and run the Preparation step again (If you already defined the samples when you ran the preparation the first time, you can skip this).
After the samples are prepared, you need to check the config files that all settings are correctly set. As for the hybrid validation sample, mostly all options are marked by a _validation and shown here:
| Setting | Type | Explanation |
|---|---|---|
samples_validation |
dict |
Same as the samples_training option but for when you want to produce the hybrid validation sample. Here you need to add the validation samples which will be used for the resampled hybrid validation file. THE ORDER OF THE PROCESSES/FLAVOURS NEEDS TO BE CONSISTENT WITH THE samples_training! |
custom_n_jets_initial |
dict or None |
These values are used only in the count and weighting method! Do not set these for the pdf method! For the other methods, these are the used jets per sample to ensure a smooth hybrid sample of t\bar{t} and Z'. We need to define some empirically derived values for the t\bar{t} samples. |
n_jets_validation |
int |
The number of jets in the final hybrid validation file. |
outfile_name_validation |
str |
Name of the output file of the hybrid validation resampling. The output file will have this name plus _resampled. |
intermediate_index_file_validation |
str |
For the resampling, the indices of the jets to use are saved in an intermediate indices .h5 file. You can define a name and path in the Preprocessing-parameters.yaml. |
DO NOT USE THIS FILES FOR TRAINING
This preprocessing config example should only be used to create the hybrid validation samples. This example should be adapted to reflect your training sample resampling method.
After this is all set, you an simple run the following command:
preprocessing.py --config <path to config file> --resampling --hybrid_validation